The bacterium Pseudomonas aeruginosa is almost everywhere — in the soil, water, and vegetation. While it does not cause disease for most people, it can lead to serious disease or death for people with compromised immune systems.
The bacteria easily adhere to surfaces, creating structures called biofilms. Joshua Shrout, professor in the Department of Civil and Environmental Engineering and Earth Sciences in the College of Engineering, and collaborators have discovered that these biofilms are not the same throughout, and that substances called alkyl quinolones (AQs) are critical to this diversity.
“We didn’t know how diverse the bacterial behavior was in such a small amount of space,” said Shrout, whose article was published in the Journal of Bacteriology. “Understanding that if you treat it as homogenous, and it’s not, is one of the reasons classic antibiotics can fail: These bacteria are literally not behaving the same here as they are over there.”
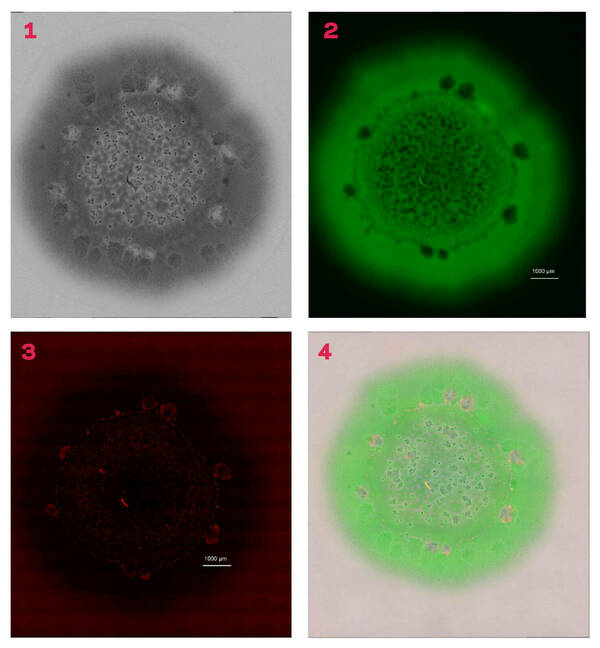
The research team began their study with what they thought was the simplest example of bacterial growth: a colony of cells on a petri plate. “It turns out this canonical example of microbiology isn’t basic or simple at all,” said Shrout, who is affiliated with the Integrated Imaging Facility and the Berthiaume Institute of Precision Health.
Lead author Abigail Weaver, assistant research professor in the Department of Civil and Environmental Engineering and Earth Sciences, first noticed the presence or absence of large-scale structures within these bacterial colonies.
“The imaging that is used here is lightyears beyond what we could have done a decade ago, which is why no one has seen this diversity before,” Shrout said.
Weaver coordinated use of light microscopy and chemical imaging to collect a series of high-resolution pictures. When reassembled, the profiles showed unexpected variety within these colony biofilms — a variety that occurs behaviorally as well as spatially. This approach allows researchers to obtain different information about these biofilms with each layer they reassemble.
Pointing to an image of a biofilm, Shrout described areas of green, which are the live cells, and said that the image was overlaid with a standard light image. In addition to the cells, there are visible places, when viewed under high magnification, where there are gaps in the biofilm. To determine whether those blank areas have dead cells or simply did not have cells, researchers overlaid another layer that displayed the dead cells in red.
“In most of the cases where it looked like cells were missing, there were actually dead cells,” Shrout said. “It seems that they killed themselves off.”
Graduate students Jin Jia and Allison R. Cutri, in the laboratory of Paul Bohn, the Arthur J. Schmitt Professor in the Department of Chemistry and Biochemistry, used chemical imaging to confirm the presence of AQs in the same areas where dead cells were present.
To further understand the heterogeneous nature of the biofilm, the researchers combined high magnification light and chemical imaging to match up spatial features with chemical features. They determined four distinct regions in these colony biofilms that are differentiated by the presence or absences of large-scale structures, and studied the importance of how AQs are associated with aggregation formation and cell death.

The AQs made by the bacteria seemed critical to making the diverse areas of the biofilms, so to test that hypothesis, researchers deleted one gene in the Pseudomonas aeruginosa that prevented that bacteria from making any AQs.
“We still see some differences in spatial heterogeneity with the AQ-deficient strain. But we don't see any of these pockets, where we see these aggregates or huge gaps of cells missing, right? It just looks pretty clean,” said Shrout, who is also a concurrent professor in the Department of Biological Sciences in the College of Science, as he showed the images of the biofilm after gene deletion. “So that allows us to pretty definitively conclude these AQs are important for when we see all this crazy heterogeneity, including the aggregates and the dead cells.”
Though researchers knew AQs were important to the development of biofilms, the new knowledge about the extensive diversity within the biofilm, in such a small space, might aid in either the development or application of new drug classes to treat Pseudomonas aeruginosa.
“If you’re treating this infection as if it’s homogeneous, and it’s not, then you don’t get the result you expect. But you shouldn’t be surprised, right?” Shrout said. “Clearly, one of the reasons that classic antibiotics can fail is that these bacteria are literally not behaving the same here as over there.”
In addition to Shrout, Weaver, and Bohn, other researchers on the paper include Jin Jia, Allison R. Cutri, Chinedu Madukoma, and Catherine Vaerewyck.
This research was funded by the National Institute of Allergy and Infectious Diseases and the Indiana Clinical and Translational Sciences Institute.
Originally published by at science.nd.edu on June 04, 2024.